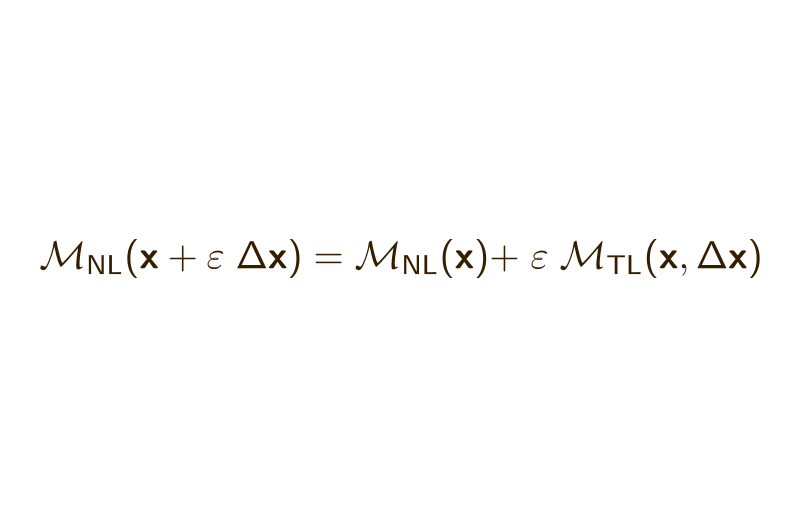Articles
Multiple biotic interactions establish phytoplankton community structure across environmental gradients
Abstract: The combination of taxa and size classes of phytoplankton that coexist at any location affects the structure of the marine food web and the magnitude of carbon fluxes to the deep ocean. But what controls the patterns of this community structure across environmental gradients remains unclear. Here, we focus on the North East Pacific Transition Zone, a ≈10° region of latitude straddling warm, nutrient‐poor subtropical and cold, nutrient‐rich subpolar gyres. Data from three cruises to the region revealed intricate patterns of phytoplankton community structure: poleward increases in the number of cell size classes; increasing biomass of picoeukaryotes and diatoms; decreases in diazotrophs and Prochlorococcus ; and both increases and decreases in Synechococcus . These patterns can only be partially explained by existing theories. Using data, theory, and numerical simulations, we show that the patterns of plankton distributions across the transition zone are the result of gradients in nutrient supply rates, which control a range of complex biotic interactions. We examine how interactions such as size‐specific grazing, multiple trophic strategies, shared grazing between several phytoplankton size classes and heterotrophic bacteria, and competition for multiple resources can individually explain aspects of the observed community structure. However, it is the combination of all these interactions together that is needed to explain the bulk compositional patterns in phytoplankton across the North East Pacific Transition Zone. The synthesis of multiple mechanisms is essential for us to begin to understand the shaping of community structure over large environmental gradients.
Modelling considerations for research on ocean alkalinity enhancement (OAE)
Abstract: The deliberate increase of ocean alkalinity (referred to as Ocean Alkalinity Enhancement or OAE) has been proposed as a method for removing CO2 from the atmosphere. Before OAE can be implemented safely, efficiently, and at scale several research questions have to be addressed including: 1) which alkaline feedstocks are best suited and in what doses can they be added safely, 2) how can net carbon uptake be measured and verified, and 3) what are the potential ecosystem impacts. These research questions cannot be addressed by direct observation alone but will require skillful and fit-for-purpose models. This chapter provides an overview of the most relevant modeling tools, including turbulence-, regional- and global-scale biogeochemical models, and techniques including approaches for model validation, data assimilation, and uncertainty estimation. Typical biogeochemical model assumptions and their limitations are discussed in the context of OAE research, which leads to an identification of further development needs to make models more applicable to OAE research questions. A description of typical steps in model validation is followed by proposed minimum criteria for what constitutes a model that is fit for its intended purpose. After providing an overview of approaches for sound integration of models and observations via data assimilation, the application of Observing System Simulation Experiments (OSSEs) for observing system design is described within the context of OAE research. Criteria for model validation and intercomparison studies are presented. The article concludes with a summary of recommendations and potential pitfalls to be avoided.
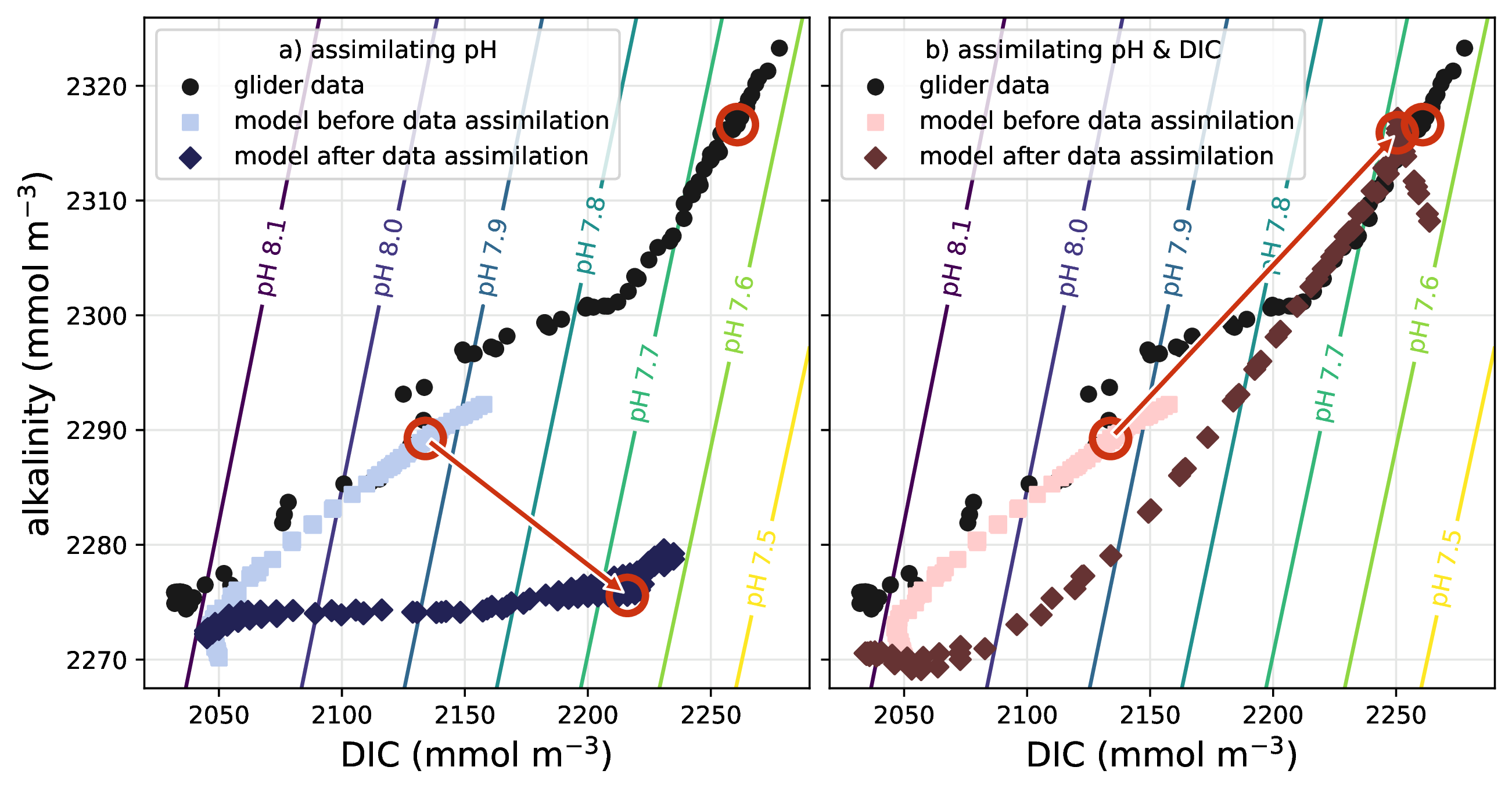
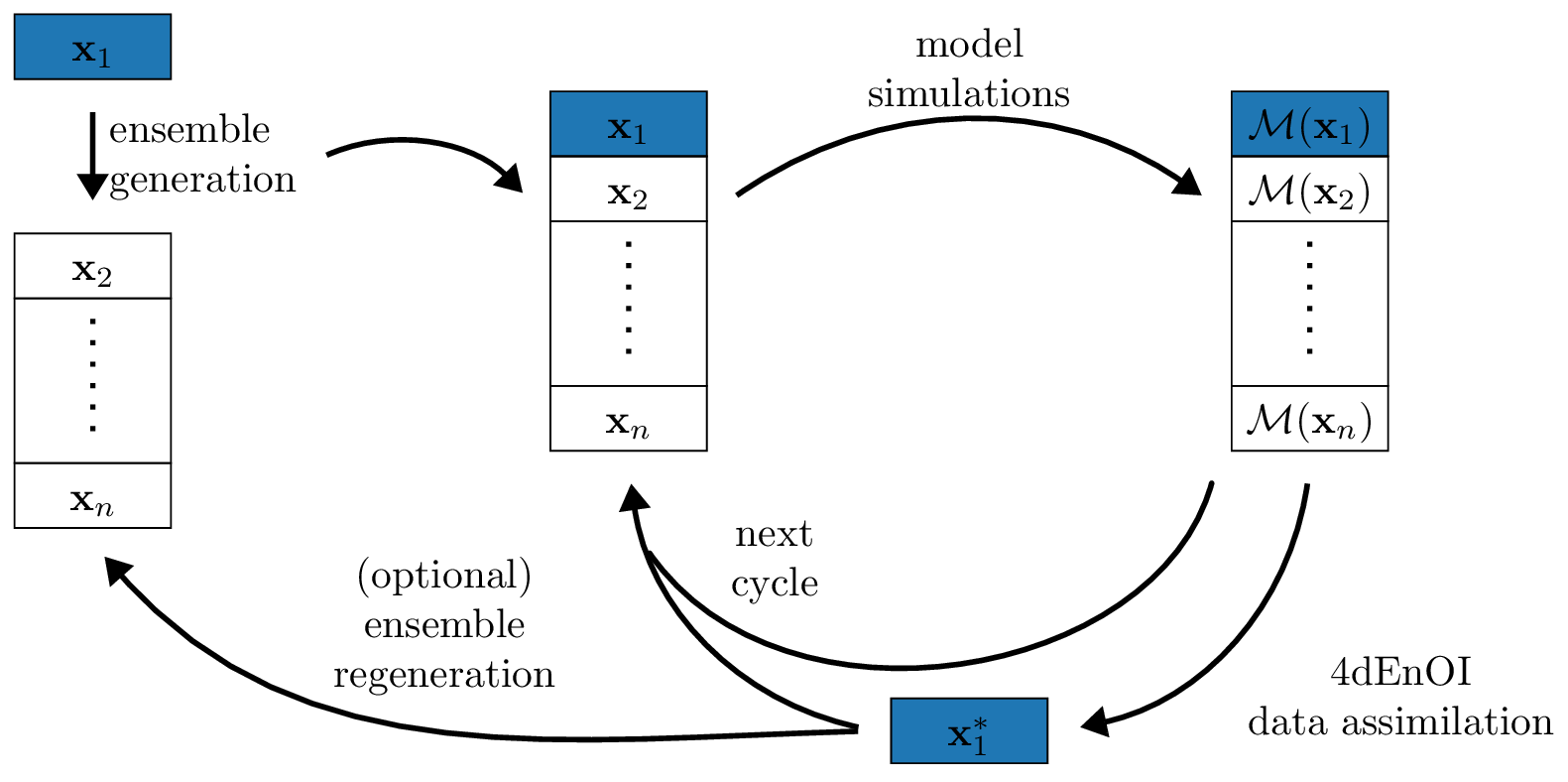
Ensemble optimal interpolation for adjoint-free biogeochemical data assimilation
Abstract: Advanced marine ecosystem models can contain more than 100 biogeochemical variables, making data assimilation for these models a challenging prospect. Traditional variational data assimilation techniques like 4dVar rely on tangent linear and adjoint code, which can be difficult to create for complex ecosystem models with more than a few dozen variables. More recent hybrid ensemble-variational data assimilation techniques use ensembles of model forecasts to produce model statistics and can thus avoid the need for tangent linear or adjoint code. We present a new implementation of a four-dimensional ensemble optimal interpolation (4dEnOI) technique for use with coupled physical-ecosystem models. Our 4dEnOI implementation uses a small ensemble, and spatial and variable covariance localization to create reliable flow-dependent statistics. The technique is easy to implement, requires no tangent linear or adjoint code, and is computationally suitable for advanced ecosystem models. We test the 4dEnOI implementation in comparison to a 4dVar technique for a simple marine ecosystem model with 4 biogeochemical variables, coupled to a physical circulation model for the California Current System. In these tests, our 4dEnOI reference implementation performs similarly well to the 4dVar benchmark in lowering the model observation misfit. We show that the 4dEnOI results depend heavily on covariance localization generally, and benefit from variable localization in particular, when it is applied to reduce the coupling strength between the physical and biogeochemical model and the biogeochemical variables. The 4dEnOI results can be further improved by small modifications to the algorithm, such as multiple 4dEnOI iterations, albeit at additional computational cost.
Evaluating Alternate Methods of 4D-Var Data Assimilation in a Coupled Hydrodynamic—Four-Component Biogeochemical Model of the California Current System
Abstract: Assimilating biogeochemical (BGC) data into ocean models using traditional four-dimensional variational data assimilation (4D-Var) includes the technical challenge of constructing tangent-linear (TLM) and adjoint (ADJ) models corresponding to the non-linear BGC model. This hurdle can be time-consuming, particularly for non-linear BGC models experiencing active development, with regular updates to functional types or representation of key BGC processes. We evaluate two alternate approaches that greatly simplify TLM and ADJ construction and eliminate the need for code updates when the underlying, non-linear BGC model changes. One form of model-reduced data assimilation represents BGC interactions as a linear combination of orthogonal modes; we use empirical orthogonal functions and refer to this as the BioEOFs method. The second approach treats BGC variables as passive tracers that experience advection and diffusion by TLM and ADJ physics over an assimilation cycle, but are biochemically inactive. We evaluate the efficacy of these methods in a realistically configured, data-assimilative implementation of the Regional Ocean Modeling System with a simple nutrient–phytoplankton–zooplankton–detritus (NPZD) BGC model for the U.S. west coast. In a series of sequential data-assimilative model cycles, observations of temperature, salinity, sea-level anomaly, and phytoplankton biomass constrain the modeled state over the period spanning January–June 2019. We perform the assimilation in both physical and logarithmic-transformed space and compare results of the two approximate methods to traditional 4D-Var using TLM and ADJ models with BGC processes corresponding explicitly to the BGC model. While the full-adjoint 4D-Var results in the best correction to phytoplankton fields for analyses and forecasts, the BioEOFs and passive-tracer approaches also significantly reduce the model-observation misfit compared to a non-assimilative run and do so at reduced computational expense. The passive tracer performs generally better than BioEOFs, whose corrections exhibit large-scale structure across the domain. While the most accurate state estimate will result from development and application of the full TLM and ADJ, the log-transformed passive-tracer approach may be a viable alternative for performing BGC 4D-Var for state estimation when a full-adjoint option is not yet available.
Ocean biogeochemical modelling
Abstract: Ocean biogeochemical models describe the ocean’s circulation, physical properties, biogeochemical properties and their transformations using coupled differential equations. Numerically approximating these equations enables simulation of the dynamic evolution of the ocean state in realistic global or regional spatial domains, across time spans from years to centuries. This Primer explains the process of model construction and the main characteristics, advantages and drawbacks of different model types, from the simplest nutrient–phytoplankton–zooplankton–detritus model to the complex biogeochemical models used in Earth system modelling and climate prediction. Commonly used metrics for model-data comparison are described, alongside a discussion of how models can be informed by observations via parameter optimization or state estimation, the two main methods of data assimilation. Examples illustrate how these models are used for various practical applications, ranging from carbon accounting, ocean acidification, ocean deoxygenation and fisheries to observing system design. Access points are provided, enabling readers to engage in biogeochemical modelling through practical code examples and a comprehensive list of publicly available models and observational data sets. Recommendations are given for best practices in model archiving. Lastly, current limitations and anticipated future developments and challenges of the models are discussed.
A Bayesian approach to modeling phytoplankton population dynamics from size distribution time series
Abstract: The rates of cell growth, division, and carbon loss of microbial populations are key parameters for understanding how organisms interact with their environment and how they contribute to the carbon cycle. However, the invasive nature of current analytical methods has hindered efforts to reliably quantify these parameters. In recent years, size-structured matrix population models (MPMs) have gained popularity for estimating division rates of microbial populations by mechanistically describing changes in microbial cell size distributions over time. Motivated by the mechanistic structure of these models, we employ a Bayesian approach to extend size-structured MPMs to capture additional biological processes describing the dynamics of a marine phytoplankton population over the day-night cycle. Our Bayesian framework is able to take prior scientific knowledge into account and generate biologically interpretable results. Using data from an exponentially growing laboratory culture of the cyanobacterium Prochlorococcus , we isolate respiratory and exudative carbon losses as critical parameters for the modeling of their population dynamics. The results suggest that this modeling framework can provide deeper insights into microbial population dynamics provided by size distribution time-series data.
Dual number-based variational data assimilation: Constructing exact tangent linear and adjoint code from nonlinear model evaluations
Abstract: Dual numbers allow for automatic, exact evaluation of the numerical derivative of high-dimensional functions at an arbitrary point with minimal coding effort. We use dual numbers to construct tangent linear and adjoint model code for a biogeochemical ocean model and apply it to a variational (4D-Var) data assimilation system when coupled to a realistic physical ocean circulation model with existing data assimilation capabilities. The resulting data assimilation system takes modestly longer to run than its hand-coded equivalent but is considerably easier to implement and updates automatically when modifications are made to the biogeochemical model, thus making its maintenance with code changes trivial.
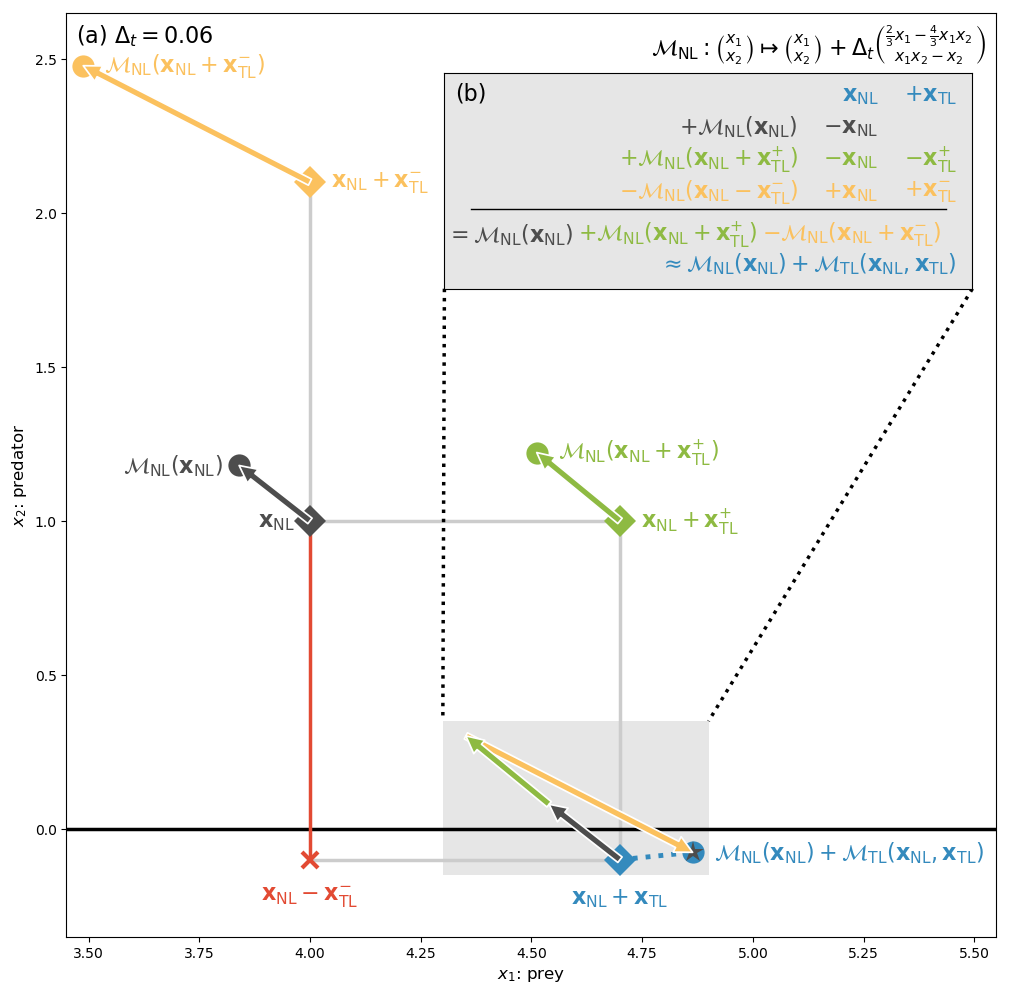
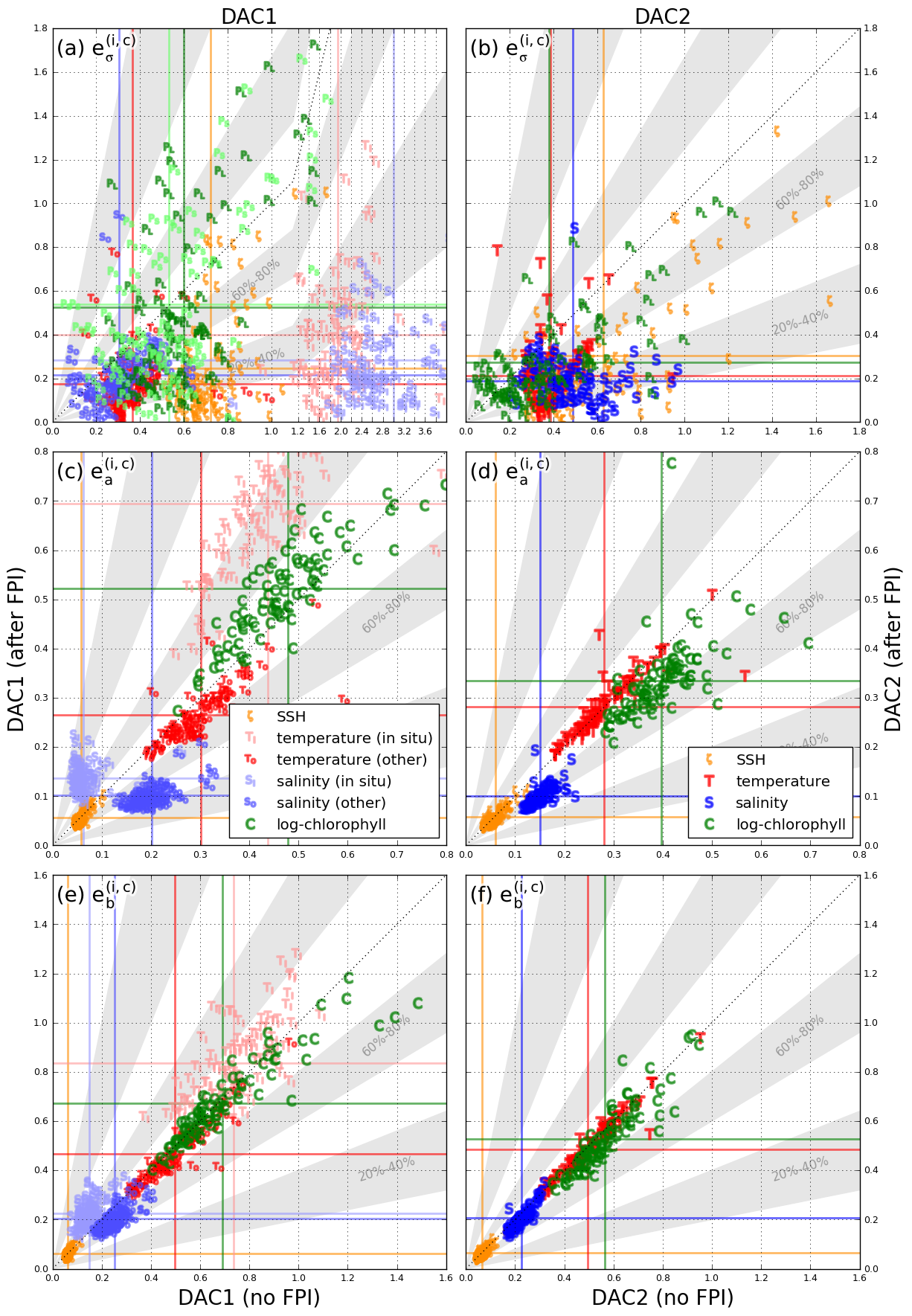
A Simple Finite Difference-Based Approximation for Biogeochemical Tangent Linear and Adjoint Models
Abstract: We present a technique that accurately approximates tangent linear and adjoint models for data assimilation applications using only evaluations of the nonlinear model. The approximation offers a simple way to create tangent linear and adjoint model code that is easily maintainable, as only major changes to the nonlinear model formulation necessitate modifications of the tangent linear or adjoint model code. The approach is particularly well‐suited to marine biogeochemical models and takes advantage of typical features of these types of models to be computationally viable. We illustrate the approximation in a realistic application, using a 3‐dimensional coupled physical‐biogeochemical 4D‐Var data assimilation system, set in the California Current system, in which the approximation is only applied to the 11 state variable biogeochemical model. In this application, the approximation‐based model solution tracks the reference solution accurately over 30 4‐day assimilation cycles but leads to a 3%-15% increase in the computational cost compared to the hand‐coded reference.
Modeling of nitrogen and phosphorus profiles in sediment of Osaka Bay , Japan with parameter optimization using the polynomial chaos expansion
Abstract: Coastal sediments adjacent to urban centers often receive high loads of organic matter (OM) due to large nutrient inputs from land that stimulate algae blooms. Early diagenetic models describing the remineralization of this OM in sediments have been developed for 50 years. Although these models can be applied to a range of marine sediments, specifying their model parameter values is difficult. In this study, one of the early diagnetic models was applied to simulate sediments in Osaka Bay, Japan and the polynomial chaos expansion (PCE) technique was used in order to choose optimal model parameters in the model. Following a sensitivity analysis, we estimated values for six parameters including the ratio of fast-decaying OM to total OM, the ratio of non-degradable OM to total OM, and the carbon–nitrogen ratio. Optimal parameter values were determined by minimizing the misfits between simulated and observed release rates of ammonium and phosphate from the sediments, and vertical profiles of inorganic nitrogen, and phosphorus in the porewater. Simulations with the optimized parameters successfully reduce a dimensionless root mean square error by 68% and agree better with the observed profiles and release rates than without parameter estimation.
Improving Variational Data Assimilation through Background and Observation Error Adjustments
Abstract: A procedure to objectively adjust the error covariance matrices of a variational data assimilation system is presented. It is based on popular diagnostics that utilize differences between observations and prior and posterior model solutions at the observation locations. In the application to a data assimilation system that combines a three-dimensional, physical-biogeochemical ocean model with large datasets of physical and chlorophyll a observations, the tuning procedure leads to a decrease in the posterior model-observation misfit and small improvements in short-term forecasting skill. It also increases the consistency of the data assimilation system with respect to diagnostics, based on linear estimation theory, and reduces signs of overfitting. The tuning procedure is easy to implement and only relies on information that is either prescribed to the data assimilation system or can be obtained from a series of short data assimilation experiments. The implementation includes a lognormal representation for biogeochemical variables and associated modifications to the diagnostics. Furthermore, the effect of the length of the observation window (number and distribution of observations) used to compute the diagnostics and the effect of neglecting model dynamics in the tuning procedure are examined.
Data assimilation of physical and chlorophyll a observations in the California Current System using two biogeochemical models
Abstract: Biogeochemical numerical models coupled to physical ocean circulation models are commonly combined with data assimilation in order to improve the models' state or parameter estimates. Yet much still needs to be learned about important aspects of biogeochemical data assimilation, such as the effect of model complexity and the importance of more realistic model formulations on assimilation results. In this study, 4D-Var-based state estimation is applied to two biogeochemical ocean models: a simple NPZD model with 4 biogeochemical variables (including 1 phytoplankton, 1 zooplankton) and the more complex NEMURO model, containing 11 biogeochemical variables (including 2 phytoplankton, 3 zooplankton). Both models are coupled to a 3-dimensional physical ocean circulation model of the U.S. west coast based on the Re- gional Ocean Modelling System (ROMS). Chlorophyll satellite observations and physical observations are assimilated into the model, yielding substantial improvements in state estimates for the observed physi- cal and biogeochemical variables in both model formulations. In comparison to the simpler NPZD model, NEMURO shows a better overall fit to the observations. The assimilation also results in small improve- ments for simulated nitrate concentrations in both models and no apparent degradation of the output for other unobserved variables. The forecasting skill of the biogeochemical models is strongly linked to model performance without data assimilation: for both models, the improved fit obtained through assimilation degrades at similar relative rates, but drops to different absolute levels. Despite the better performance of NEMURO in our experiments, the choice of model and desired level of complexity should depend on the model application and the data available for assimilation.
Simple parameter estimation for complex models — Testing evolutionary techniques on 3-dimensional biogeochemical ocean models
Abstract: Parameter estimation is an important part of numerical modeling and often required when a coupled physical–biogeochemical ocean model is first deployed. However, 3-dimensional ocean model simulations are computationally expensive and models typically contain upwards of 10 parameters suitable for estimation. Hence, manual parameter tuning can be lengthy and cumbersome. Here, we present four easy to implement and flexible parameter estimation techniques and apply them to two 3-dimensional biogeochemical models of different complexities. Based on a Monte Carlo experiment, we first develop a cost function measuring the model-observation misfit based on multiple data types. The parameter estimation techniques are then applied and yield a substantial cost reduction over ∼ 100 simulations. Based on the outcome of multiple replicate experiments, they perform on average better than random, uninformed parameter search but performance declines when more than 40 parameters are estimated together. Our results emphasize the complex cost function structure for biogeochemical parameters and highlight dependencies between different parameters as well as different cost function formulations.
Model investigations of the North Atlantic spring bloom initiation
Abstract: The spring bloom - a massive growth of phytoplankton that occurs annually during the spring season in mid and high latitudes - plays an important role in carbon export to the deep ocean. The onset of this event has been explained from bottom-up and top-down perspectives, exemplified by the ‘‘critical-depth'' and the ‘‘dilution-recoupling'' hypotheses, respectively. Both approaches differ in their key expectations about how seasonal fluctuations of the mixed layer affect the plankton community. Here we assess whether the assumptions inherent to these hypotheses are met inside a typical onedimensional Nutrient-Phytoplankton-Zooplankton-Detritus (NPZD) model, optimized to best represent climatological annual cycles of satellite-based phytoplankton biomass in the Subpolar North Atlantic. The optimized model is used in idealized experiments that isolate the effects of mixed layer fluctuations and zooplankton grazing, in order to elucidate their significance. We analyzed the model sensitivity qualitatively and using a second-order Taylor series decomposition of the model equations. Our results show that the conceptual bases of both bottom-up and top-down approaches are required to explain the process of blooming; however, neither of their bloom initiation mechanisms fully applies in the experiments. We find that a spring bloom can develop in the absence of mixed layer fluctuations, and both its magnitude and timing seem to strongly depend on nutrient and light availability. Furthermore, although zooplankton populations modulate the phytoplankton concentrations throughout the year, directly prescribed and physically driven changes in zooplankton grazing do not produce significant time shifts in bloom initiation, as hypothesized. While recognizing its limitations, our study emphasizes the processes that require further testing in order to discern among competing hypotheses.
Periodic time-dependent parameters improving forecasting abilities of biological ocean models
Abstract: Using two emulator-based procedures, we estimate time-dependent values for two key plankton parameters in a three-dimensional biogeochemical (BGC) ocean model. The estimation is based on a 4 year time series of daily surface satellite chlorophyll observations. The estimated parameters display an annual periodicity that can be explained by the succession of plankton groups in the study region. Model simulations using these parameters show improved fit to observations and better forecasting abilities compared to simulations with constant optimal parameters; the newly introduced sequential parameter estimation procedure creates the strongest improvement. The inclusion of time-dependent parameters represents a simple way to improve the predictive skill of BGC models and their representation of plankton dynamics.
Simulating sediment-water exchange of nutrients and oxygen: A comparative assessment of models against mesocosm observations
Abstract: How to represent nutrient fluxes resulting from organic matter remineralization in sediments should be an important consideration when formulating a biogeochemical ocean model. Here representations ranging from simple parameterizations to vertically resolved diagenetic models are compared against a comprehensive, multi-year data set from a mesocosm eutrophication study. Observations of sediment-water fluxes of nutrients and oxygen and measurements of the state of the overlying water column were made over 2.5 years in nine mesocosms, six of which received geometrically increasing loads of inorganic nutrients. These observations are used here to force and optimize two simple parameterizations of sediment oxygen uptake, one representative two-layer diagenetic model and one representative multi-layer diagenetic model. In cross-validation experiments the predictive ability of these different representations is compared. The main results are that the optimized multi-layer model fits the observations best and also proved to be the most parsimonious, while the two-layer model failed the cross-validation indicating that it is prone to over-fitting and was less parsimonious even than one of the simpler functional oxygen flux models. We recommend that sediment models that are candidates for inclusion in a biogeochemical model be assessed through a process of optimization and cross-validation as we have done here. {\textcopyright} 2013 Elsevier Ltd.
Particle filter-based data assimilation for a three-dimensional biological ocean model and satellite observations
Abstract: We assimilate satellite observations of surface chlorophyll into a three-dimensional biological ocean model in order to improve its state estimates using a particle filter referred to as sequential importance resampling (SIR). Particle Filters represent an alternative to other, more commonly used ensemble-based state estimation techniques like the ensemble Kalman filter (EnKF). Unlike the EnKF, Particle Filters do not require normality assumptions about the model error structure and are thus suitable for highly nonlinear applications. However, their application in oceanographic contexts is typically hampered by the high dimensionality of the model’s state space. We apply SIR to a high-dimensional model with a small ensemble size (20) and modify the standard SIR procedure to avoid complications posed by the high dimensionality of the model state. Two extensions to the SIR include a simple smoother to deal with outliers in the observations, and state- augmentation which provides the SIR with parameter memory. Our goal is to test the feasibility of biological state estimation with SIR for realistic models. For this purpose we compare the SIR results to a model simulation with optimal parameters with respect to the same set of observations. By running replicates of our main experiments, we assess the robustness of our SIR implementation. We show that SIR is suitable for satellite data assimilation into biological models and that both extensions, the smoother and state- augmentation, are required for robust results and improved fit to the observations.
Sensitivity and uncertainty analysis of model hypoxia estimates for the Texas-Louisiana shelf
Abstract: Numerical ocean models are becoming increasingly important tools for marine research and for management of marine resources. It is therefore crucial that uncertainty in model predictions and model sensitivity to errors in the model inputs be quantified. We performed a combined sensitivity and uncertainty analysis for a realistic physical-biological model of the Texas-Louisiana shelf in the northern Gulf of Mexico. The model simulates the major physical and biological processes involved in the formation of the hypoxic zone that develops on the shelf every summer. With the help of a statistical emulator technique, we introduced uncertainty in selected model inputs and assessed the effects of these uncertainties on the predicted development and spatial distribution of bottom hypoxia. The uncertain inputs we examined belong to two categories: (i) biological inputs including river nutrient concentration, phytoplankton growth rate and initial and boundary conditions of biological variables, and (ii) physical inputs including freshwater river runoff, wind forcing, and mixing coefficients. We show that uncertainty in different inputs has distinct effects on model output which vary in magnitude, time, and space. Uncertainty in physical inputs was found to have a strong impact on estimates of hypoxia, e.g., hypoxic area estimates vary by more than 40%, due to a 20% variation in the freshwater river runoff.
Estimating time-dependent parameters for a biological ocean model using an emulator approach
Abstract: We use a statistical emulator technique, the polynomial chaos expansion, to estimate time-dependent values for two parameters of a 3-dimensional biological ocean model. We obtain values for the phytoplankton carbon-to-chlorophyll ratio and the zooplankton grazing rate by minimizing the misfit between simulated and satellite-based surface chlorophyll. The misfit is measured by a spatially averaged, time-dependent dis- tance function. A cross-validation experiment demonstrates that the influence of outlying satellite data can be diminished by smoothing the distance function in time. The optimal values of the two parameters based on the smoothed distance function exhibit a strong time-dependence with distinct seasonal differences, without overfitting observations. Using these time-dependent parameters, we derive (hindcast) state esti- mates in two distinct ways: (1) by using the emulator-based interpolation and (2) by performing model runs with time-dependent parameters. Both approaches yield chlorophyll state estimates that agree better with the observations than model estimates with globally optimal, constant parameters. Moreover, the em- ulator approach provides us with estimates ofparameter-induced model state uncertainty, which help deter- mine at what time improvement in the model simulation is possible. The time-dependence of the analyzed parameters can be motivated biologically by naturally occurring seasonal changes in the composition of the plankton community. Our results suggest that the parameter values of typical biological ocean models should be treated as time-dependent and will result in a better representation of plankton dynamics in these models. We further demonstrate that emulator techniques are valuable tools for data assimilation and for analyzing and improving biological ocean models.
Data assimilation with a local Ensemble Kalman Filter applied to a three-dimensional biological model of the Middle Atlantic Bight
Abstract: A multivariate sequential data assimilation approach, the Localized Ensemble Kalman Filter (LEnKF), was used to assimilate daily satellite observations of ocean chlorophyll into a three-dimensional physical–biolog- ical model of the Middle Atlantic Bight (MAB) for the year 2006. Covariance localization was applied to make the EnKF analysis more effective by removing spurious long-range correlations in the ensemble approxima- tion of the model’s covariance. The model is based on the Regional Ocean Modeling System (ROMS) and coupled to a biological nitrogen cycle model, which includes seven state variables: chlorophyll, phytoplank- ton, nitrate, ammonium, small and large detrital nitrogen, and zooplankton. An ensemble of 20 model simu- lations, generated by perturbing the biological parameters according to assumed probability distributions, was used. Model fields of chlorophyll, phytoplankton, nitrate and zooplankton were updated at all vertical layers during LEnKF analysis steps, based on their cross-correlations with surface chlorophyll (the observed variable). The performance of the LEnKF scheme, its influence on the model’s predictive skill and on surface particulate organic matter concentrations and primary production are investigated. Estimates of surface chlo- rophyll and particulate organic carbon are improved in the data-assimilative simulation when compared to one without any assimilation, as is the model’s predictive skill.
A geologically constrained Monte Carlo approach to modeling exposure ages from profiles of cosmogenic nuclides: An example from Lees Ferry, Arizona
Abstract: We present a user‐friendly and versatile Monte Carlo simulator for modeling profiles of in situ terrestrial cosmogenic nuclides (TCNs). Our program (available online at http://geochronology.earthsciences.dal. ca/downloads‐models.html) permits the incorporation of site‐specific geologic knowledge to calculate most probable values for exposure age, erosion rate, and inherited nuclide concentration while providing a rigorous treatment of their uncertainties. The simulator is demonstrated with 10Be data from a fluvial terrace at Lees Ferry, Arizona.
Introduction and Assessment of Measures for Quantitative Model-Data Comparison Using Satellite Images
Abstract: Satellite observations of the oceans have great potential to improve the quality and predictive power of numerical ocean models and are frequently used in model skill assessment as well as data assimilation. In this study we introduce and compare various measures for the quantitative comparison of satellite images and model output that have not been used in this context before. We devised a series of test to compare their performance, including their sensitivity to noise and missing values, which are ubiquitous in satellite images. Our results show that two of our adapted measures, the Adapted Gray Block distance and the entropic distance D2, perform better than the commonly used root mean square error and image correlation.
Sequential data assimilation applied to a physical-biological model for the Bermuda Atlantic time series station
Abstract: In this study, we investigate sequential data assimilation approaches for state estimation and prediction in a coupled physical–biological model for the Bermuda Atlantic Time Series (BATS) site. The model is 1- dimensional (vertical) in space and based on the General Ocean Turbulence Model (GOTM). Coupled to GOTM is a biological model that includes phytoplankton, detritus, dissolved inorganic nitrogen, chlorophyll and oxygen. We performed model ensemble runs by introducing variations in the biological parameters, each of which was assigned a probability distribution. We compare and contrast here 2 sequential data assimilation methods: the ensemble Kalman filter (EnKF) and sequential importance resampling (SIR). We assimilated different types of BATS observations, including particulate organic nitrogen, nitrate+nitrite, chlorophyll a and oxygen for the 2-year period from January 1990 to December 1991, and quantified the impact of the data assimilation on the model’s predictive skill. By applying a cross-validation to the data- assimilative and deterministic simulations we found that the predictive skill was improved for 2-week forecasts. In our experiments the EnKF, which exhibited a stronger effect on the ensemble during the assimilation step, showed slightly higher improvements in the predictive skill than the SIR, which preserves dynamical model consistency in our implementation. Our numerical experiments show that statistical properties stabilize for ensemble sizes of 20 or greater with little improvement for larger ensembles.